Research: Project 1
 Succinate production by Actinobacillus succinogenes. If produced fermentatively at a price competitive with that of petroleum derivative
maleic anhydride, succinate could replace maleic anhydride as the precursor of bulk
chemicals, which represent a $15 billion per year commodity chemical market. The Gram-negative
rumen bacterium A. succinogenes is the best natural succinate producer known, but it also produces acetate, formate,
and ethanol as byproducts. Our goal is to engineer A. succinogenes to efficiently produce succinate as the sole fermentation product.
Succinate production by Actinobacillus succinogenes. If produced fermentatively at a price competitive with that of petroleum derivative
maleic anhydride, succinate could replace maleic anhydride as the precursor of bulk
chemicals, which represent a $15 billion per year commodity chemical market. The Gram-negative
rumen bacterium A. succinogenes is the best natural succinate producer known, but it also produces acetate, formate,
and ethanol as byproducts. Our goal is to engineer A. succinogenes to efficiently produce succinate as the sole fermentation product.
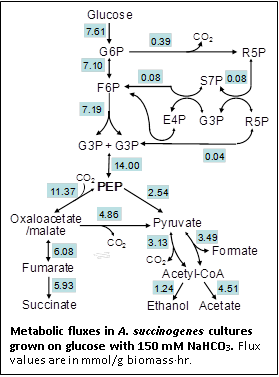
Our 13C-based metabolic flux analysis (MFA) studies showed that during A. succinogenes growth on glucose, most of the NADPH needed for growth is generated by transhydrogenase activity or by malate decarboxylation through malic enzyme. Significant forward and reverse fluxes through malic enzyme and/or oxaloacetate decarboxylase indicate that phosphoenolpyruvate, oxaloacetate, malate, and pyruvate represent four nodes for flux distribution between succinate and other fermentation products. High NaHCO3 and H2 concentrations increase succinate production at the expense of acetate and formate production. They do so not by increasing the carboxylation flux from phosphoenolpyruvate to oxaloacetate, but by decreasing the decarboxylating flux from malate and/or oxaloacetate to pyruvate and by increasing the pyruvate carboxylating flux. Pyruvate and formate dehydrogenase fluxes vary in response to the different redox demands imposed by the different NaHCO3 and H2 concentrations. These metabolic constraints should be taken into consideration to design effective metabolic engineering strategies for increased succinate production.
To allow us to engineer A. succinogenes for enhanced succinate production, we constructed an A. succinogenes-Escherichia coli shuttle plasmid vector, and we developed a markerless knockout method based on the E. coli isocitrate dehydrogenase gene (icd) as the selection marker.
Current work for this project involves (i) characterizing the mutations identified in A. succinogenes strains evolved for fast growth on xylose and other lignocellulose sugars; (ii) optimizing A. succinogenes microaerobic growth on glycerol, a byproduct of biodiesel production; (iii) engineering A. succinogenes for anaerobic growth on glycerol; (iv) characterizing the metabolism of A. succinogenes on xylose and glycerol by MFA; and (v) engineering A. succinogenes for homosuccinate fermentation.