Going up: Discovering a protein’s ‘elevator motion’ could spur new cancer treatments
Michigan State University researcher Jian Hu has taken another important step in learning as much as possible about tiny protein machines that help shuttle metals into living cells.
This latest step, published in the journal Nature Communications, provides detailed new insights into how these machines work. Though this is a study in fundamental biology, Hu and his team are working to use this knowledge to develop new cancer therapies and enable people to live healthier lives.
Organisms across the tree of life — including bacteria, plants and people — use these proteins, which are called ZIPs for short. That stands for “Zrt-/Irt-like proteins,” which looks a little intimidating, but essentially means that these proteins transport zinc, iron and manganese, in most cases.
Although there’s only a tiny bit of these metals in biological systems, ZIPs are critically important for healthy living.

Part of the research team at Michigan State University that has revealed how the protein family known as ZIP works in atomic detail. From left to right: graduate students Peixuan Yu (rotation student) and Yuhan Jiang, postdoctoral research accociate Yao Zhang, Associate Professor Jian Hu and MSU Foundation Professor Guowei Wei.
A more intimate understanding of how ZIPs transport these metals through cell membranes would better equip researchers to study dysfunctions caused by problematic mutations. But even when the proteins are functioning properly, they present important health considerations.
For example, in humans, many types of cancer cells host an abnormally large number of a protein called ZIP4.
“There are 14 different types of ZIPs in humans that are involved in different biological functions,” said Hu. “We’re particularly interested in ZIP4 because it’s aberrantly upregulated in about half of the different types of cancer, including breast cancer, ovarian cancer and pancreatic cancer.”
Additionally, a ZIP in plants that transports life-essential iron is also implicated in absorbing cadmium from soil. Cadmium is a toxic metal found in industrial pollution and contaminated soils.
“That’s what we’re working toward, but we need to understand their fundamental properties and behaviors first,” Hu said.
With support from the National Institutes of Health and the Plant Resilience Institute at MSU, Hu and his colleagues focused on some of that fundamental science in their new report. The researchers provided the first comprehensive explanation for how the proteins physically move metals into cells.
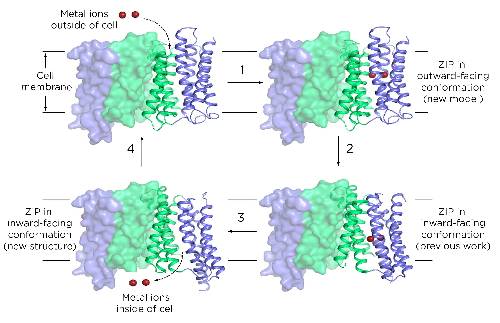
What the team found was that ZIPs have internal structures that move up and down to transport metals.
“This is called the elevator-type mode,” Hu said. His team was the first to propose this transport mode, initially at the 65th Biophysical Society Annual Meeting and now by publishing it in a peer-reviewed journal.
Hu stressed that this discovery was a team effort, with collaborators providing valuable insights from different research backgrounds.
“This work provides structural, computational, biochemical and functional evidence to convincingly support the proposed transport mechanism,” Hu said. “Previously, people had no idea how this works.”
The team included Guowei Wei, an MSU Foundation Professor in the College of Natural Science, and Min Su, who was an assistant research scientist at the University of Michigan during this project. He’s now the director of the Center for Electron Microscopy at the University of Missouri.
Molecular biology is a team sport
When it comes to understanding proteins, knowing what they look like is a big deal. It helps researchers see how the biomolecules work and is a prerequisite to applications such as designing drugs that can target the protein.
When it comes to ZIPs, though, determining their structure is also incredibly challenging.
ZIPs were first discovered in the 1990s, but it wasn’t until 2017 that researchers knew with certainty what one looked like. That was thanks to Hu and his team publishing research on a ZIP found in bacteria.
“That was great. It was the first time we saw the structure,” Hu said. “But it didn’t solve the problem of how these things work.”
Researchers knew that ZIPs had to change their conformation to collect metals from outside a cell and then release them into the cell’s interior. The 2017 paper was a milestone, showing what one of those conformations looked like for the first time. Yet a single structure goes only so far in explaining a dynamic process. It was akin to having a still frame from a movie.
In the new paper, researchers have filled in the rest of that movie. For their part, Hu and members of his lab discovered a new frame. That is, they’ve shown a ZIP structure in a different conformation than what they published in 2017.
They’ve also been working with collaborators who could help them make the most of that new structure, in part, through computational modeling led by Guowei Wei’s research group.
“This is a significant achievement and we’re really proud of it,” said Wei, an MSU Foundation Professor in the Department of Mathematics and the Department of Biochemistry and Molecular Biology. “It shows the power of collaboration between experiment and simulation.”
For its new report, the team now had two experimentally determined structures and a computational model that researchers validated with biochemical studies. Together, these set important constraints on how a ZIP could reorient itself as it ferried metals from point A to point B. With those constraints, the team figured out how different parts of the protein could shift as the ZIP passed metals through a cell membrane.

The research team led by Michigan State University also included (top to bottom) Kaifu Gao and Dexin Sui, formerly of MSU; and Min Su, who is the director of the University of Missouri’s Electron Microscopy Core. Gao now works in industry as a senior machine learning drug discovery scientist and Sui is a research laboratory specialist at the Life Sciences Institute at the University of Michigan.
Although making this breakthrough took six years, the researchers are optimistic that the pace of progress will accelerate with their new discovery. With more data available, researchers can start thinking about how to study ZIPs with more tools, including artificial intelligence.
During the pandemic, for example, Wei and his team published several papers in the span of months, using AI to make predictions about the novel coronavirus, such as which variants would become dominant.
“For the coronavirus, there were thousands of labs producing data simultaneously,” Wei said. “For ZIPs, there wasn’t that much data available. Dr. Hu is one of the few people focusing on these proteins.”
Now, Hu, Wei and Hideki Takahashi, an associate professor of biochemistry and molecular biology at MSU, are working to secure funding for a project that would apply machine learning to studies of plant ZIPs.
“It takes time at the beginning,” said Wei, “but I believe this is going to enable a lot of things and help the future impact grow."
This story originally appeared on the MSU Today website.