Freeze frame: Study uses novel sequencing method to pinpoint toxic changes in liver cells

Imagine being able to look at individual frozen liver cells and determine if a chemical, drug, or
supplement was toxic by changes in the cells’ ribonucleic acid, or RNA, levels.
In a recent paper titled “Single nuclei RNA sequencing assessment of the hepatic effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin,” published in the journal Cellular and Molecular Gastroenterology and Hepatology, that’s exactly what researchers at Michigan State University have shown may be feasible.
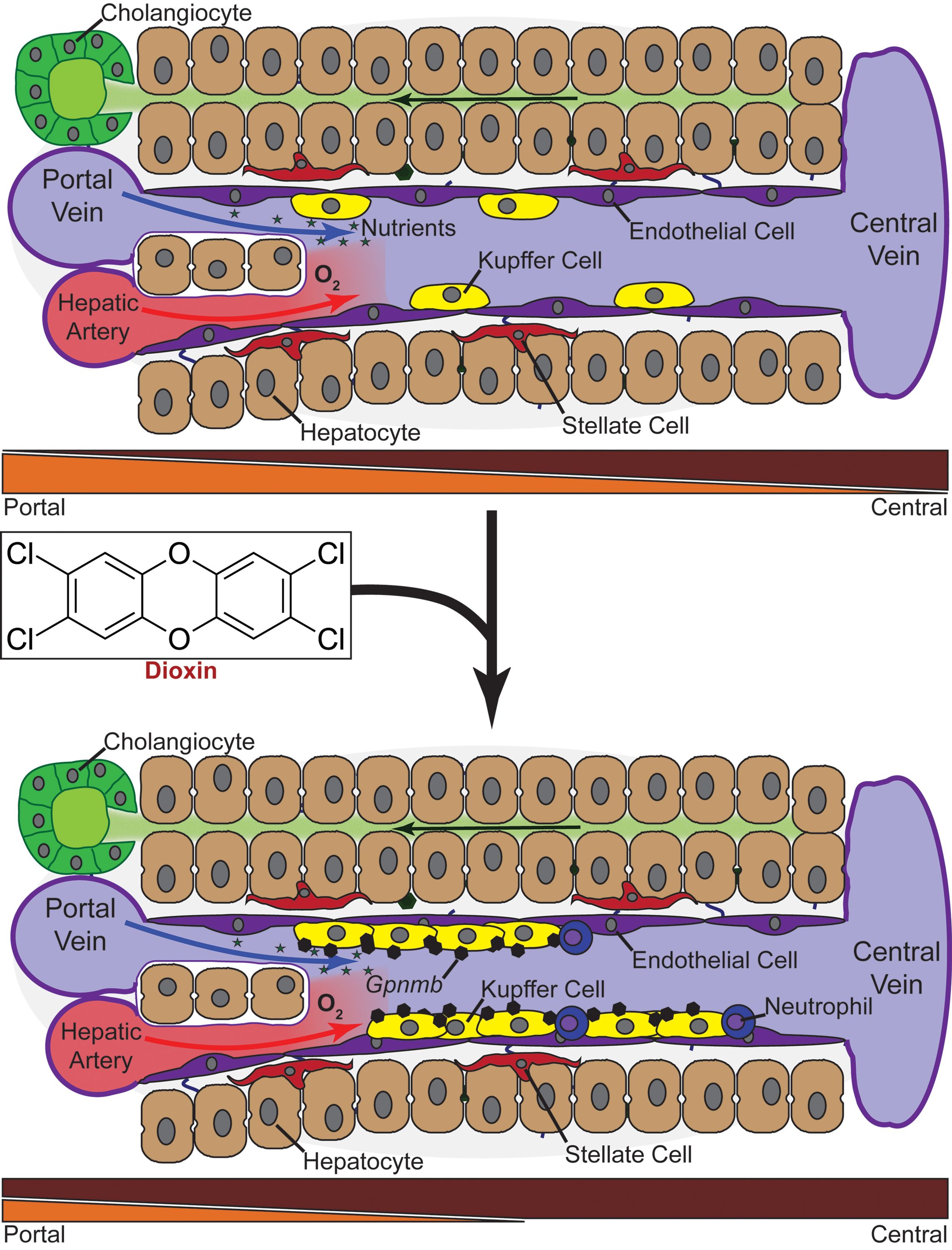
The study, led by MSU Department of Biochemistry and Molecular Biology (BMB) research associate Rance Nault, in the lab of BMB professor Tim Zacharewski, shows, for the first time, that frozen liver samples can be used for single-cell RNA sequencing to examine changes in liver cell types following exposure to dioxin—the contaminant in Agent Orange, and a pollutant found throughout the environment including Superfund sites in Michigan and the United States.
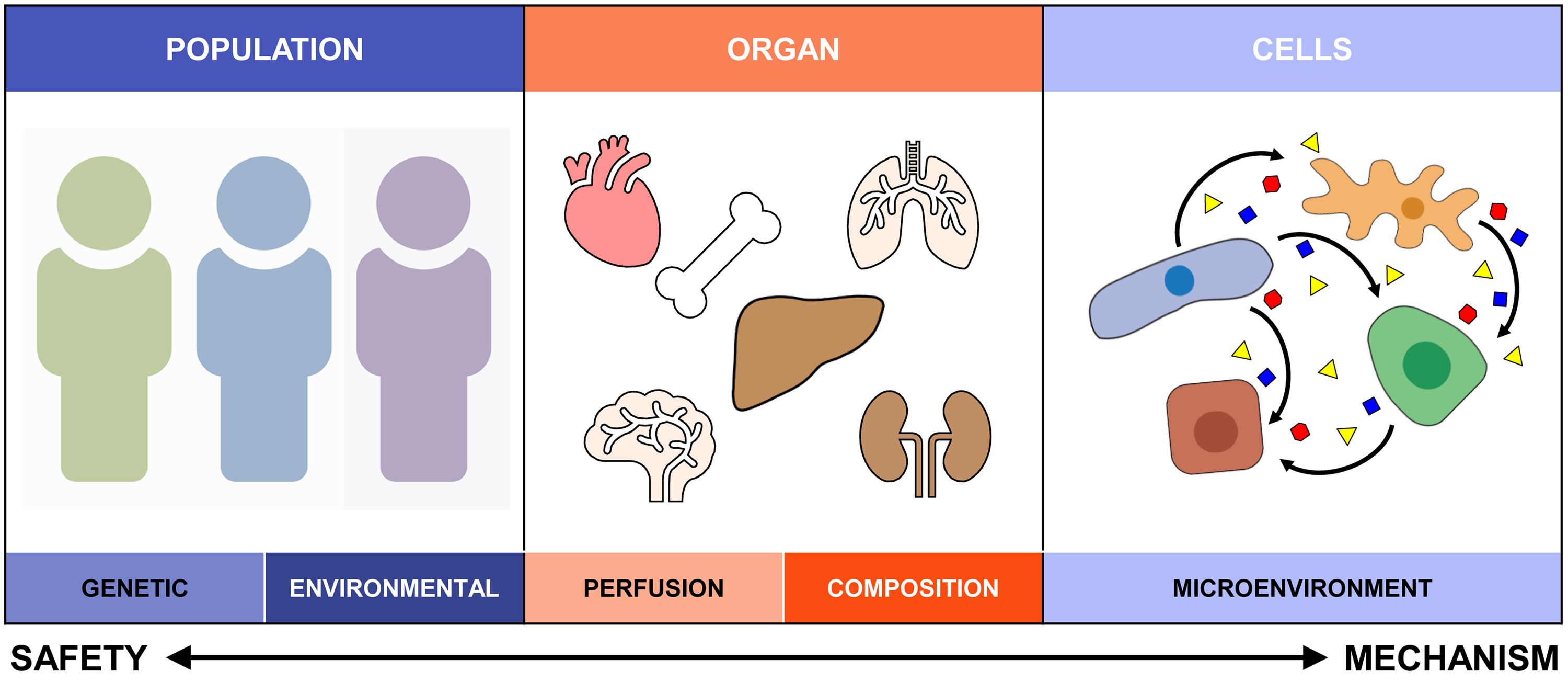
“Typically, if researchers wanted to study the toxicity of a substance, they would grind up whole pieces of tissue and measure the average levels of RNA for each gene in the sample,” Rance said. “This process, called bulk RNA sequencing, was very informative but did not consider the diversity of cell types in a tissue. For example, at least 11 different cell types are present in the liver and each one may react differently to a chemical, drug, or supplement.”
The Zacharewski lab studies the adverse effects of dioxins and related compounds—commonly found Superfund contaminants—on human health and, more specifically, their effects on liver function. Superfund sites are areas contaminated with hazardous waste and chemicals that are improperly managed and may pose a threat to human health and the environment. These sites may include abandoned manufacturing facilities, processing plants, landfills and mining sites. Currently, there are 65 Superfund sites here in Michigan.
Within the past five years, a new technology has been developed—single-cell RNA sequencing—that allows researchers to measure the levels of RNA in individual cells of a tissue sample. This technology is more precise and provides new insights about how individual cell types respond when exposed to unfamiliar and potentially toxic chemicals.
However, the technology can often be complicated by the need to use fresh, unfrozen samples could limit its usefulness for toxicology studies intended to determine the safety of drugs, chemicals, and supplements at multiple dose levels.
In this study, Nault successfully demonstrated how this technique can be used to examine frozen liver samples following exposure to a chemical that can cause significant liver toxicity, expanding its use in toxicology to support the safety assessments of environmental contaminants, drugs, and dietary/health supplements.
“This paper shows that single-cell sequencing technology is a viable option to better understand how chemicals and drugs affect liver function,” Nault said. “It’s exciting to get into this new, innovative area of research and help bring this technology to the field of toxicology.”
The study was funded by the Superfund Research Program (NIEHS SRP P42ES04911) and the National Human Genome Research Institute (NHGRI R21 HG010789).
Banner image: Mouse liver exposed to Dioxin showing the accumulation of fat in hepatocytes, the major cell type of the liver. Photo: Rance Nault



