Fishing for biochemistry’s ‘dark matter'
Article Highlights
- Aleksandra Skirycz, an associate professor in the Department of Biochemistry and Molecular
Biology, has received a five-year, $1.91 million award from the NIH to uncover the
secrets of crucial small molecules called metabolites.
- The group will target protein-metabolite interactions by casting a wide analytical 'net' into what's been called biochemistry's 'dark matter,' where vast quantities of metabolite functions aren't yet known.
- These findings will lead to fundamental discoveries linked to the health and resilience of plants and animals.
To some, it’s “dark matter.” For others, terra incognita: unexplored land.
These are just some of the phrases biochemists use when describing the metabolome, the set of all small molecules, or metabolites, in a biological sample.
More specifically, it’s a nod to the large number of chemical signatures discovered during metabolomic analysis that are unidentifiable. In fact, by most estimates, less than a few percentages of all measured compounds can be chemically annotated.
“When we talk of dark matter, we’re really talking about the functions of metabolites,” said Aleksandra Skirycz, an associate professor in Michigan State University’s Department of Biochemistry and Molecular Biology.
“So, even for the metabolites we do chemically understand, we often don’t know their roles.”
The existence of these mystery metabolites is a tantalizing target for researchers, especially those looking to better understand their impact on organismal health.
Now, with the support of a Maximizing Investigators’ Research Award, or MIRA, from the National Institutes of Health, Skirycz and her lab are breaking new ground in the quest to better define the secrets of the metabolome.
The five-year, $1.91 million award will support novel work that explores interactions between metabolites and proteins by fishing with a wider analytical “net.”
Already used as world-changing drugs and agrichemicals, identifying the roles of these small molecules will lead to fundamental discoveries linked to the health and resilience of plants and animals.
Show me your friends, and I’ll tell you who you are
While scientists have broadly examined the ways proteins interact with proteins, how proteins and metabolites interact is still a hugely understudied phenomenon.
This disparity has led the Skirycz Group to develop more creative and efficient ways to characterize the dark matter of the metabolome and identify the previously unknown functions of compounds.
“More often than not a metabolite needs to target a protein to execute its function, the same way a drug in the body must target a protein to achieve its goals,” Skirycz explained.
“We realized if we could find a way to map protein targets for the metabolites we were interested in, we’d be able to say something about their functions. Essentially, ‘show me your friends, and I’ll tell you who you are.’”
At the Max Planck Institute of Molecular Plant Physiology in Golm, Germany, Skirycz began mapping how proteins and metabolites interacted in different organisms.
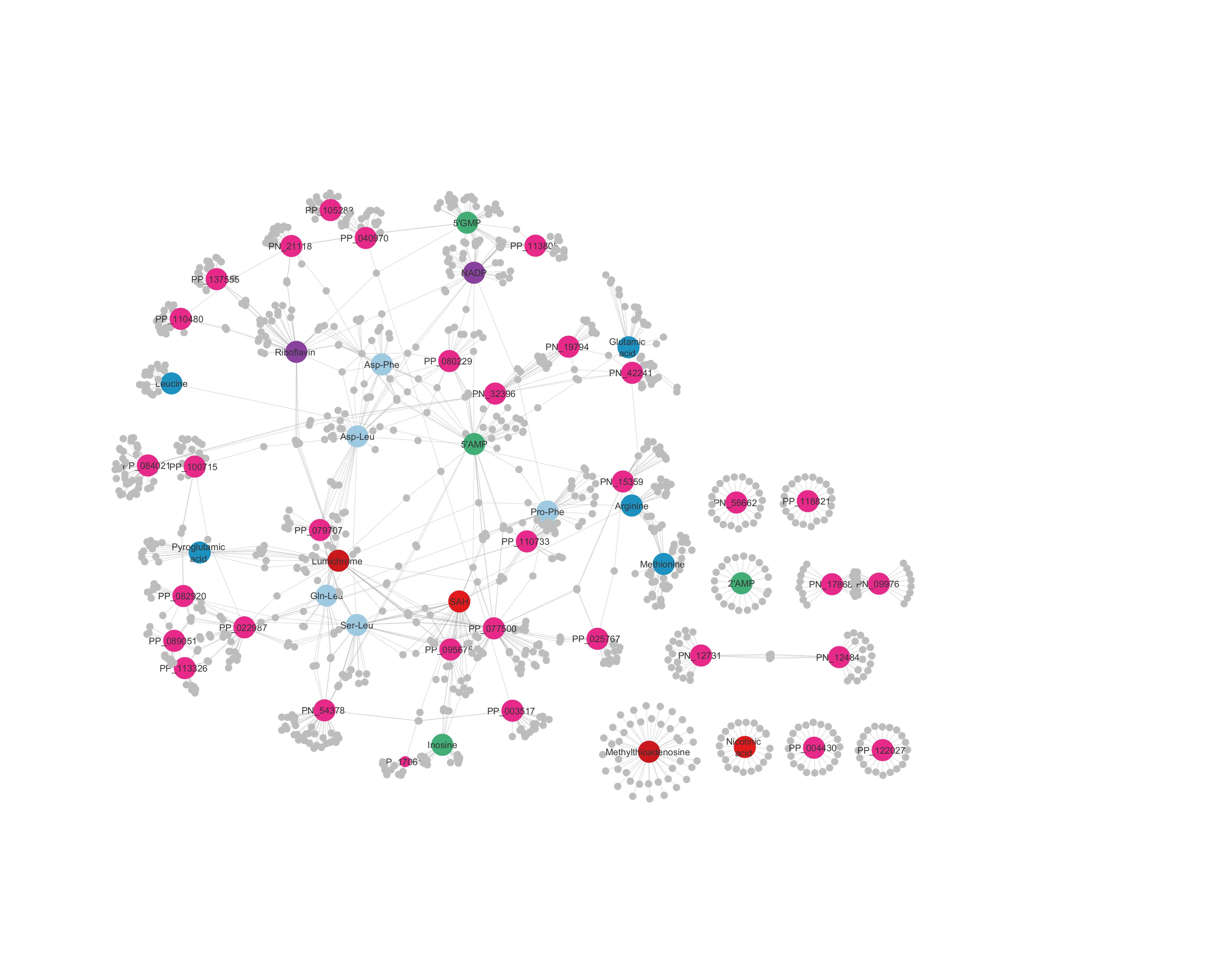
Known as interaction networks, these maps visually represent the likelihood of metabolite-protein interactions.
At the center of a map, you’ll find interactions that are well known to researchers. Move toward the edges, however, and you’ll start to find interactions that are suspected, but not yet proven. These hypothetical interactions are exciting targets for researchers to eventually validate.
“When validating edges, you want to be as clever as possible,” Skirycz said, noting the extensive time and effort required for case-by-case validation.
“The real risk comes from not knowing if you can validate something, or if it will turn out interesting and functional after all.”
Going fishing
With her MIRA, Skirycz will compare multiple interaction networks through a creative approach that seeks to overcome some of these analytical challenges — one that leverages MSU’s cutting-edge research facilities.
“The university has a fantastic Mass Spectrometry and Metabolomics Core, as well as Proteomics Core. This was an important factor for joining MSU,” Skirycz said, who moved her lab to MSU in early 2024.
Specifically, Skirycz will compare the interaction networks of four model organisms: nematode C. elagans, yeast S.cerevisiae, bacteria E. coli, and the plant A. thaliana.
If the team can identify pairings that occur across multiple networks and in familiar ways — for instance, metabolites interacting with proteins with similar structure — these can be flagged as significant and worth validating further.
Since they’re found in multiple organisms, there’s also a higher chance these pairings are relevant to general organismal health.
To accomplish their analysis, the researchers will use mass spectrometry to measure metabolites and proteins using what’s called an untargeted approach.
Describing different methods of analysis, Skirycz offered fishing as a useful metaphor.
In a targeted approach, biochemists can ‘fish’ with a particular metabolite as bait, hoping to attract and reel in certain proteins. Proteins might also be used as bait for specific metabolites.
An untargeted approach, by contrast, is closer to using a net that pulls in a diverse and sometimes surprising catch.
“We don’t have to define what we want to measure at the start, and can instead measure a larger portfolio. Often, we’ll find a small molecule that we love and can then validate with a more targeted method,” Skirycz explained.

“To increase precision, it’s always a combination of approaches.”
The overall process the Skirycz Group has developed is called PROMIS, which stands for PROtein-Metabolite Interactions using Size separation.
This process builds off a form of chemical analysis called co-fractionation/mass spectrometry, which is highly suited for looking at interactions at a global scale.
By separating a cell’s protein-metabolite complexes based on size and analyzing how these elements naturally group together, the researchers can discover hints that point to possible interactions worthy of investigation.
Ultimately, the group’s MIRA will help shine a further light on metabolomic dark matter and the vast scientific potential of undiscovered metabolite functions that could lead to healthier, more resilient plants and animals.
Reflecting on her time in the Amazon while at the ITV Institute of Technology in Belem, Brazil, Skirycz noted the exciting outcomes of working with the unknown.
“I realized we knew so little about the function of these molecules and their vast chemical diversity. These compounds aren’t produced by the organisms to please us,” she said.
“If we know that these molecules act by interacting with certain proteins, we can speed up identification of interesting and beneficial compounds.”



