Current Research
Plants utilize inorganic nutrients in the environment to produce essential metabolites. Nutrient use efficiency thus affects plant fitness and yield performance. Acquisition of nitrogen (N) and sulfur (S) is particularly important since these elements are required for synthesizing amino acids and proteins. Nitrate (NO3-), ammonium (NH4+) and sulfate (SO42-) are the major forms of N and S utilized by plants in nature. However, these nutrient resources are not uniformly distributed in the soil environment. Despite the application of fertilizers, the soil nutrient concentrations fluctuate due to climate changes.
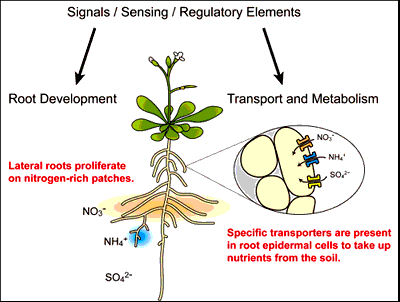
My research interests are focused on molecular mechanisms of metabolic and morphological strategies that plant roots have developed as genetic traits to control N and S assimilation and thus optimize their growth in the environment. Research projects in my laboratory employ multi-disciplinary approaches of plant biochemistry, molecular biology, molecular genetics and functional genomics, aiming to identify sensing and signaling components associated with regulation of transport and metabolism of N and S, and modification of root system architecture in response to changes in plant nutritional status and environmental N and S conditions.
Plant roots express specific forms of ion transporters and enzymes that facilitate N and S uptake and utilization. Abundance and catalytic efficiency of transporters and enzymes that are responsible for acquisition of N and S resources are controlled in a demand-driven manner. Significance of such response behavior has been demonstrated in S utilization mechanisms. Genes encoding sulfate transporters that facilitate sulfate uptake across the plasma membranes and sulfate unloading from vacuoles are abundantly expressed in roots under S-deficient conditions [1,2]. ATP sulfurylase, the first committing enzyme in sulfate assimilation, is post-transcriptionally regulated by microRNA-395 and dually localized in chloroplast and cytosol in an isoform-specific manner [3,4]. A transcription factor SULFUR LIMITATION1 (SLIM1) plays a key role in inducing expression of sulfate transporters and microRNA-395, while repressing glucosinolate biosynthesis in Arabidopsis under S deficiency [3,5]. Thus, the regulatory pathways for controlling sulfate transport, sulfate assimilation and secondary S metabolism are highly coordinated under control of S-responsive signals [1].
Plants can modify their root system architecture in response to changes in nutrient conditions. The morphological plasticity of the root system allows them to make efficient access to nutrient resources unevenly distributed in the soil environment. Root lengths and degrees of complexity of root branches may change depending on forms and amount of nutrients present in soil patches. N has profound effects on such mechanisms. Lateral roots proliferate on N-rich patches [6], however a contrasting root morphological phenotype can be observed under N-deficient conditions [7]. Our recent study on small signaling peptides encoded by the CLE gene family in Arabidopsis demonstrates the importance of a morphological control mechanism that prevents expansion of the root system in unfavorable N nutritional environments [7].
Plant roots acclimate to nutrient-deficient or excess conditions by coordinating the functions of transporters and enzymes. Morphological signals play important roles in shaping the root system architecture to be adjusted to varying nutrient conditions. My research group is studying these fundamental questions to gain mechanistic insights into genetic traits associated with control of N and S metabolism and plant root functions. Improvement of nutrient use efficiency is critical to development of sustainable agricultural systems. Biotechnological research and development are expected for designing crop breeding and nutrient management strategies aimed for better production of food and energy resources.
References
[1] Takahashi H, Kopriva S, Giordano M, Saito K, Hell R. (2011) Sulfur assimilation
in photosynthetic organisms: molecular functions and regulations of transporters and
assimilatory enzymes. Annu Rev Plant Biol 62, 157-184.
[2] Kataoka T, Watanabe-Takahashi A, Hayashi N, Ohnishi M, Mimura T, Buchner P, Hawkesford
MJ, Yamaya T, Takahashi H. (2004) Vacuolar sulfate transporters are essential determinants
controlling internal distribution of sulfate in Arabidopsis. Plant Cell 16, 2693-2704.
[3] Kawashima CG, Yoshimoto N. Maruyama-Nakashita A, Tsuchiya YN, Saito K, Takahashi
H. Dalmay T. (2009). Sulphur starvation induces the expression of microRNA-395 and
one of its target genes but in different cell types. Plant J 57, 313–321.
[4] Bohrer AS, Yoshimoto N, Sekiguchi A, Rykulski N, Saito K, Takahashi H. (2015).
Alternative translational initiation of ATP sulfurylase underlying dual localization
of sulfate assimilation pathways in plastids and cytosol in Arabidopsis thaliana. Front Plant Sci 5, 750. doi: 10.3389/fpls.2014.00750
[5] Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H. (2006) Arabidopsis
SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism.
Plant Cell 18, 3235-3251.
[6] Lima JE, Kojima S, Takahashi H, von Wirén N. (2010) Ammonium triggers lateral
root branching in Arabidopsis in an AMT1;3-dependent manner. Plant Cell 22, 3621-3633.
[7] Araya T, Miyamoto M, Wibowo J, Suzuki A, Kojima S, Tsuchiya YN, Sawa S, Fukuda
H, von Wirén N, Takahashi H. (2014) CLE-CLAVATA1 peptide-receptor signaling module
regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc Natl Acad Sci USA 111, 2029-2034.