Current Research
(Research Description PDF - 825 kb)
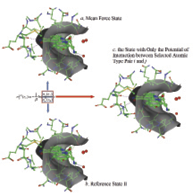
(using PDBID 1xbc) of how the KECSA
statistical potential is modeled. The
protein binding site is shown as a grey
surface with the ligand located within
the binding site surrounded by protein
residues which it makes contacts with.
The pink dashed lines indicate interactions
between certain atom pair types i and j,
(i.e. carbonyl oxygen with amine nitrogens
in this example) which are defined as
“selected interactions” in this manuscript.
Green dashed lines indicate all other
non-covalent interactions between the
protein and ligand atoms in the binding
pocket, defined as “background interactions”.
(a) In the mean force state, the system is filled
with all types of interactions. (b) The reference
state II contains all the background interactions.
(c) Removing all the background interactions
from total interactions results in a state with
only the selected interactions for each
i and j combination.
Research at the interface between the computational sciences and biology is our group focus. We work on a number of problems and collaborate with experimentalists at every opportunity. Research areas of most interest include computer-aided drug design (CADD), the role potential function error plays in drug design and protein folding, metalloenzymes and metal ion homeostasis, development and application of linear-scaling quantum mechanical methods to biological problems and NMR and X-ray structure refinement using quantum mechanical methods. For further details go to the group web page at http://merzgroup.org.
In the structure-based drug design area we are interested in developing novel tools to predict the binding affinity of ligands for a given receptor. Along these lines we developed a novel knowledge-based protein-ligand scoring function that employs a new definition for the reference state (see the Figure), allowing us to relate a statistical potential to a Lennard-Jones (LJ) potential. In this way, the LJ potential parameters were generated from protein-ligand complex structural data contained in the PDB. Forty-nine types of atomic pairwise interactions were derived using this method, which we call the knowledge-based and empirical combined scoring algorithm (KECSA). Validation results illustrate that KECSA shows improved performance in all test sets when compared with other scoring methods especially in its ability to minimize the RMSE.
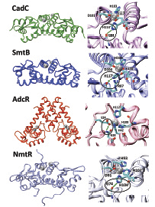
coordination in CadC (1U2W),
SmtB (1R22) AdcR (3TGN)
and NmtR (computational model)
transcriptional regulators. Zinc
ions are shown as silver spheres
and the hydrogen bonds are
shown with a green line.
Metalloenzymes carry out a myriad of biological functions and we have a long-term
interest in modeling the structure and function of proteins involving metal ion catalysis
or homeostasis. A recent publication illustrates an example of the study of transition
metal homeostasis. A metal-mediated interprotomer hydrogen bond has been implicated
in the allosteric mechanism of DNA operator binding in several metal-sensing proteins.
Using computational methods, we investigated the energetics of such zinc-mediated
interactions in members of the ArsR/SmtB family of proteins (CzrA, SmtB, CadC and
NmtR) and the MarR family zinc-uptake repressor AdcR, each of which feature similar
interactions, but in sites that differ widely in their allosteric responsiveness.
We provided novel structural insight into previously uncharacterized allosteric forms
of these proteins using computational methodologies. We find this metal-mediated interaction
to be significantly stronger (~8 kcal/mol) at functional allosteric metal binding
sites compared to a non-responsive site (CadC) and the apo-proteins. Simulations of
the apo-proteins further revealed that the high interaction energy works to overcome
the considerable disorder at these hydrogen-bonding sites and functions as a “switch”
to lock in a weak DNA-binding conformation once metal is bound. These findings suggested
a globally conserved functional role of metal-mediated second-coordination shell hydrogen
bonds at allosterically responsive sites in zinc-sensing transcription regulators.