Current Research
We are using computational methods in order to study the structure, dynamics, and
energetics of biological macromolecules such as proteins or nucleic acids. Our focus
is concentrated in two areas: 1) The modeling of large supramolecular assemblies in
atomic detail and 2) the accurate prediction of native protein structures.
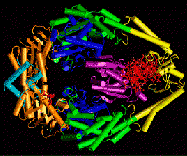
As an example of a supramolecular complex we are investigating the interaction between the E.coli mismatch recognition protein MutS and DNA with mismatched or missing base pairs. MutS recognizes defective DNA after replication and initiates a multi-step process that leads to DNA repair. A detailed understanding of the DNA mismatch repair system is relevant for some types of cancer where the repair process is compromised. Starting from crystal structures of the MutS protein-DNA complex we apply computer simulation techniques to look at energetic and dynamic aspects of this system.
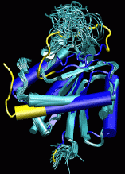 The prediction of protein structures from sequence has seen great progress through
recent methodological advances and the availability of an increasing number of structural
templates from experimental protein structures. It is now often possible to generate
approximate predictions that capture many of the general features of the native fold.
The prediction of protein structures from sequence has seen great progress through
recent methodological advances and the availability of an increasing number of structural
templates from experimental protein structures. It is now often possible to generate
approximate predictions that capture many of the general features of the native fold.
However, it remains challenging to reach levels of accuracy that are comparable to experimental data unless structures from closely related sequences are available. We are working on new methods that allow the refinement of approximate predictions towards more native-like structures by using new enhanced sampling techniques in combination with an accurate energetic description.
In both cases, the use of an implicit solvent representation is very attractive, since such a model can maintain a realistic description of a biomolecule in aqueous solution without the computational expense of explicit solvent molecules. This allows the modeling of larger systems and/or over longer time scales at a similar level of detail. We are interested in continuing the development of new implicit solvent methodologies in order to improve the level of realism in complex environments.