Current Research
Genome-wide mRNA translation and novel peptides in plant responses to the environment
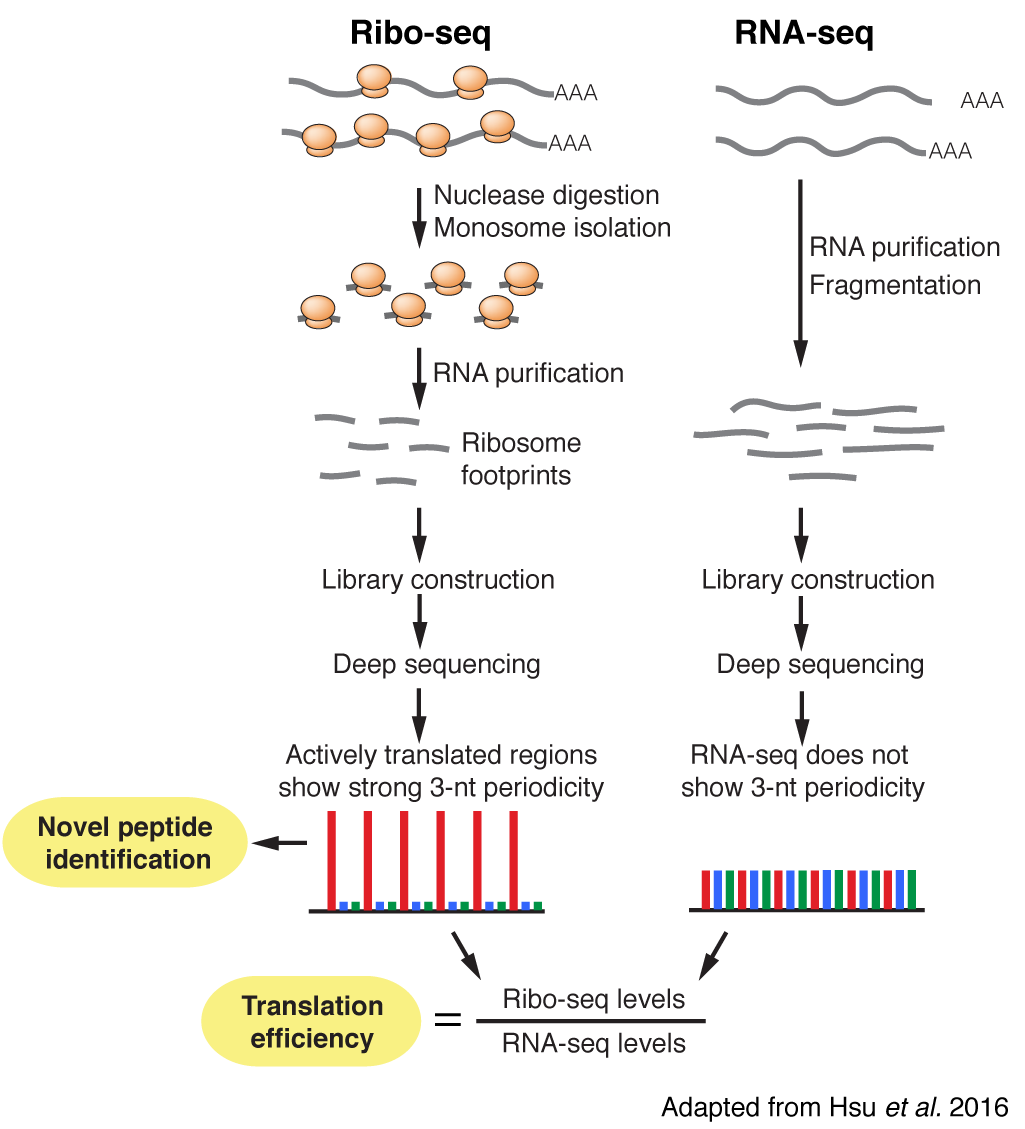
Plants are sessile organisms and have to respond rapidly to environmental changes to optimize their growth. Some of these changes, such as day/night cycles, can be predictable, while others, such as soil salt stress, are not. How plants coordinate their growth in response to these different types of changes remains unclear. Our research uses both Arabidopsis and tomato as model systems to understand how plants respond to diurnal cycles and abiotic stress. We focus on translational regulation and peptide signaling, two rapidly acting regulatory mechanisms that are poorly understood in plant science. We have established a robust approach utilizing ribosome profiling (Ribo-seq; deep sequencing of ribosome footprints) to tackle questions about translational dynamics and identify novel peptides.
1. Profile the translational landscape under diurnal cycles and in response to abiotic stress
Studies on plant responses to the environment have focused on quantifying RNA. However, translation is the next step of the central dogma and can change protein abundance more rapidly than transcription. By studying translational regulation, we seek to uncover novel mechanisms that promote immediate responses to different types of environmental stimuli.
2. Identify and characterize potential signaling peptides
Peptide signaling regulates cell-cell communication both during development and in response to the environment. There are over 600 receptor-like kinases in Arabidopsis, but most of their ligand peptides are unknown. Our strategy uses Ribo-seq combined with a computational pipeline; it is a high-throughput approach for uncovering small peptides with potential signaling activity. The functions of these new peptides are further studied using transgenic plants and genome editing tools such as CRISPR-Cas9.
3. Develop experimental and analytic tools to study translation in plants
We are actively improving the Ribo-seq technique in plants and developing visualization tools to facilitate the mining of these large-scale datasets.