Current Research
Biochemical Genomics and Genetics in the 21st Century
We are in the midst of a golden period for research into plant metabolism. The ongoing development and integration of various omics-technologies with established biochemical, molecular and genetic approaches have created a "perfect storm" in which we can computationally, genetically and biologically dissect metabolism between organisms, genomes and across evolutionary time. Historically, the most recent comparable period of advancement was during the 1960’s and 70’s, when radiolabel allowed the pathways for much of plant metabolism as we now know it to be proposed. It is quite clear we are experiencing an analogous period of incredible advancement in our fundamental understanding of plant metabolism at the individual gene, system and genome levels.
Current areas of research include:
Association Mapping for Seed Traits in Arabidopsis and Maize
 Our work in the past 10+ years focused on pioneering genomics-enabled approaches to
dissect the core tocopherol and carotenoid pathways in plants. We have at times been quite successful at pathway engineering; however, this work has also highlighted how little we understand about the integration
of plant metabolism. For example, high light stress increases leaf tocopherol levels
30-fold while our best pathway engineering effort is but 4-fold. Similarly, the seed
phenotypes of carotenoid mutants often cannot be predicted from leaf phenotypes. Such
results indicate there are processes/genes outside the core biosynthetic pathways
(e.g., transcription factors, metabolite transporters, storage and turnover pathways,
cell biology components, etc.) that contribute to desired nutritional phenotypes.
Identifying and understanding these additional loci especially in seed, the primary
plant tissue used for food, is currently and will remain a major focus of my group.
Our work in the past 10+ years focused on pioneering genomics-enabled approaches to
dissect the core tocopherol and carotenoid pathways in plants. We have at times been quite successful at pathway engineering; however, this work has also highlighted how little we understand about the integration
of plant metabolism. For example, high light stress increases leaf tocopherol levels
30-fold while our best pathway engineering effort is but 4-fold. Similarly, the seed
phenotypes of carotenoid mutants often cannot be predicted from leaf phenotypes. Such
results indicate there are processes/genes outside the core biosynthetic pathways
(e.g., transcription factors, metabolite transporters, storage and turnover pathways,
cell biology components, etc.) that contribute to desired nutritional phenotypes.
Identifying and understanding these additional loci especially in seed, the primary
plant tissue used for food, is currently and will remain a major focus of my group.
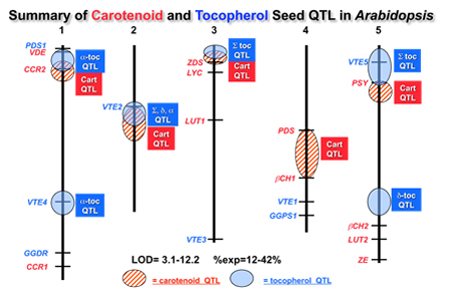
Toward this end the lab has been using quantitative genetics in Arabidopsis to identify and clone novel loci that control variation for specific micronutrients in seed. We have screened four Arabidopsis recombinant inbred populations for 12 carotenoid and 6 tocopherol seed traits. Overall, only 20% of the QTL intervals identified contain known carotenoid or tocopherol biosynthetic genes indicating that even in Arabidopsis, the majority of loci affecting two of the best-studied nutritional traits remain unknown. Identifying and understanding the action of the other 80% of the loci in Arabidopsis (and other plants) that control seed traits is a major focus of the lab.