 The Hoogstraten laboratory studies biologically and biochemically important problems
using techniques derived from biophysics and physical chemistry. Specifically, we
are interested in the relationship between conformational dynamics and molecular function
in RNA and RNA-containing systems. Current research has made it clear that an understanding
of the three-dimensional structure of proteins and nucleic acids, although incredibly
valuable, is not sufficient to provide a full picture of mechanism; instead, careful
study of how that structure changes with time, and how those fluctuations are coupled
to molecular function, is also necessary. Nuclear magnetic resonance spectroscopy
(NMR) is the key technology for the detailed, site-specific characterization of the
dynamics of biological molecules. We complement extensive NMR measurements with techniques
including studies of the thermodynamics and kinetics of macromolecular interaction
in solution using isothermal titration calorimetry (ITC) and surface plasmon resonance
(SPR), respectively; circular dichroism, fluorescence anisotropy, and other biophysical
measurements; biochemical probing of RNA enzyme kinetics and other aspects of function;
and contemporary molecular dynamics calculations.
The Hoogstraten laboratory studies biologically and biochemically important problems
using techniques derived from biophysics and physical chemistry. Specifically, we
are interested in the relationship between conformational dynamics and molecular function
in RNA and RNA-containing systems. Current research has made it clear that an understanding
of the three-dimensional structure of proteins and nucleic acids, although incredibly
valuable, is not sufficient to provide a full picture of mechanism; instead, careful
study of how that structure changes with time, and how those fluctuations are coupled
to molecular function, is also necessary. Nuclear magnetic resonance spectroscopy
(NMR) is the key technology for the detailed, site-specific characterization of the
dynamics of biological molecules. We complement extensive NMR measurements with techniques
including studies of the thermodynamics and kinetics of macromolecular interaction
in solution using isothermal titration calorimetry (ITC) and surface plasmon resonance
(SPR), respectively; circular dichroism, fluorescence anisotropy, and other biophysical
measurements; biochemical probing of RNA enzyme kinetics and other aspects of function;
and contemporary molecular dynamics calculations.
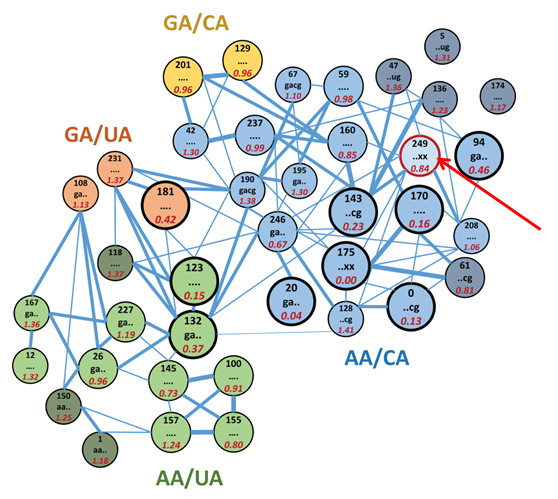
 One major area of interest is the study of the "dynamics-function" relationships underlying
catalysis by RNA enzymes (ribozymes). underlying catalysis by RNA enzymes (ribozymes).
We have developed novel technology to efficiently introduce stable isotope labels
at specific sites within the RNA ribose ring, enabling unprecedented NMR measurements
of the dynamic properties of the RNA backbone. To complement the spectroscopic technology,
we have introduced the use of chemically modified, covalently-constrained nucleotides
to test the functional relevance of backbone motions observed by NMR. Model system
work has provided novel insights into the nature of RNA tertiary structure, with the
bottom line being that RNA energy landscapes are characterized by multiple deep, narrow
potential wells, and that molecular function is often limited by the ability of the
system to dynamically sample the appropriate portions of that landscape.
One major area of interest is the study of the "dynamics-function" relationships underlying
catalysis by RNA enzymes (ribozymes). underlying catalysis by RNA enzymes (ribozymes).
We have developed novel technology to efficiently introduce stable isotope labels
at specific sites within the RNA ribose ring, enabling unprecedented NMR measurements
of the dynamic properties of the RNA backbone. To complement the spectroscopic technology,
we have introduced the use of chemically modified, covalently-constrained nucleotides
to test the functional relevance of backbone motions observed by NMR. Model system
work has provided novel insights into the nature of RNA tertiary structure, with the
bottom line being that RNA energy landscapes are characterized by multiple deep, narrow
potential wells, and that molecular function is often limited by the ability of the
system to dynamically sample the appropriate portions of that landscape.
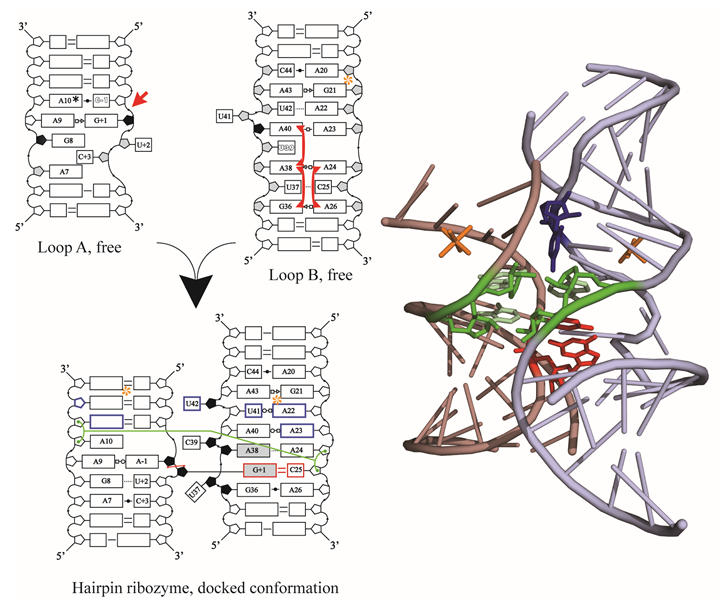 We are now applying these techniques to catalytic RNA (ribozyme) systems of interest,
including a major effort to understand the domain-docking step that underlies the
formation of catalytically active three-dimensional structure in the hairpin ribozyme.
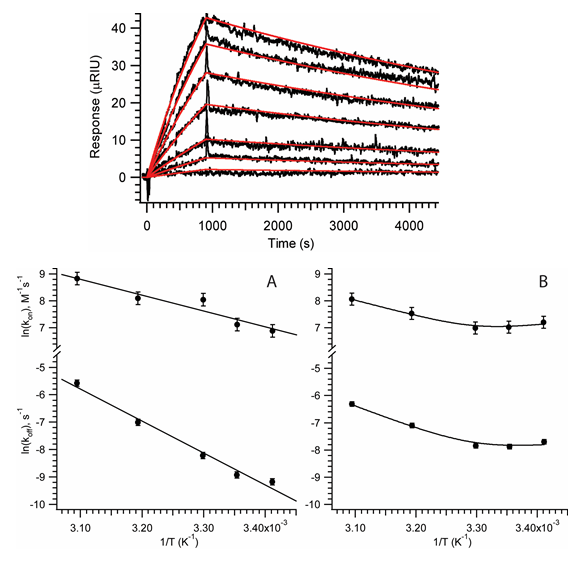
Recent advances in the hairpin system include a comprehensive biophysical characterization
of the intermolecular docking interaction using a combination of SPR analyses of docking
kinetics, a novel circular dichroism (CD) difference assay for docking equilibrium,
and enzyme kinetic data. A full analysis of the thermodynamics and kinetics of docking
has helped to reveal the mechanistic diversity of, and delicate balance of contributions
to, RNA-RNA tertiary interface formation. In collaboration with Prof. Michael Feig’s
group, we are also using state-of-the-art molecular dynamics calculations to simulate
in atomic detail the conformational sampling that underlies catalysis in this system.
We are now applying these techniques to catalytic RNA (ribozyme) systems of interest,
including a major effort to understand the domain-docking step that underlies the
formation of catalytically active three-dimensional structure in the hairpin ribozyme.
Recent advances in the hairpin system include a comprehensive biophysical characterization
of the intermolecular docking interaction using a combination of SPR analyses of docking
kinetics, a novel circular dichroism (CD) difference assay for docking equilibrium,
and enzyme kinetic data. A full analysis of the thermodynamics and kinetics of docking
has helped to reveal the mechanistic diversity of, and delicate balance of contributions
to, RNA-RNA tertiary interface formation. In collaboration with Prof. Michael Feig’s
group, we are also using state-of-the-art molecular dynamics calculations to simulate
in atomic detail the conformational sampling that underlies catalysis in this system.





