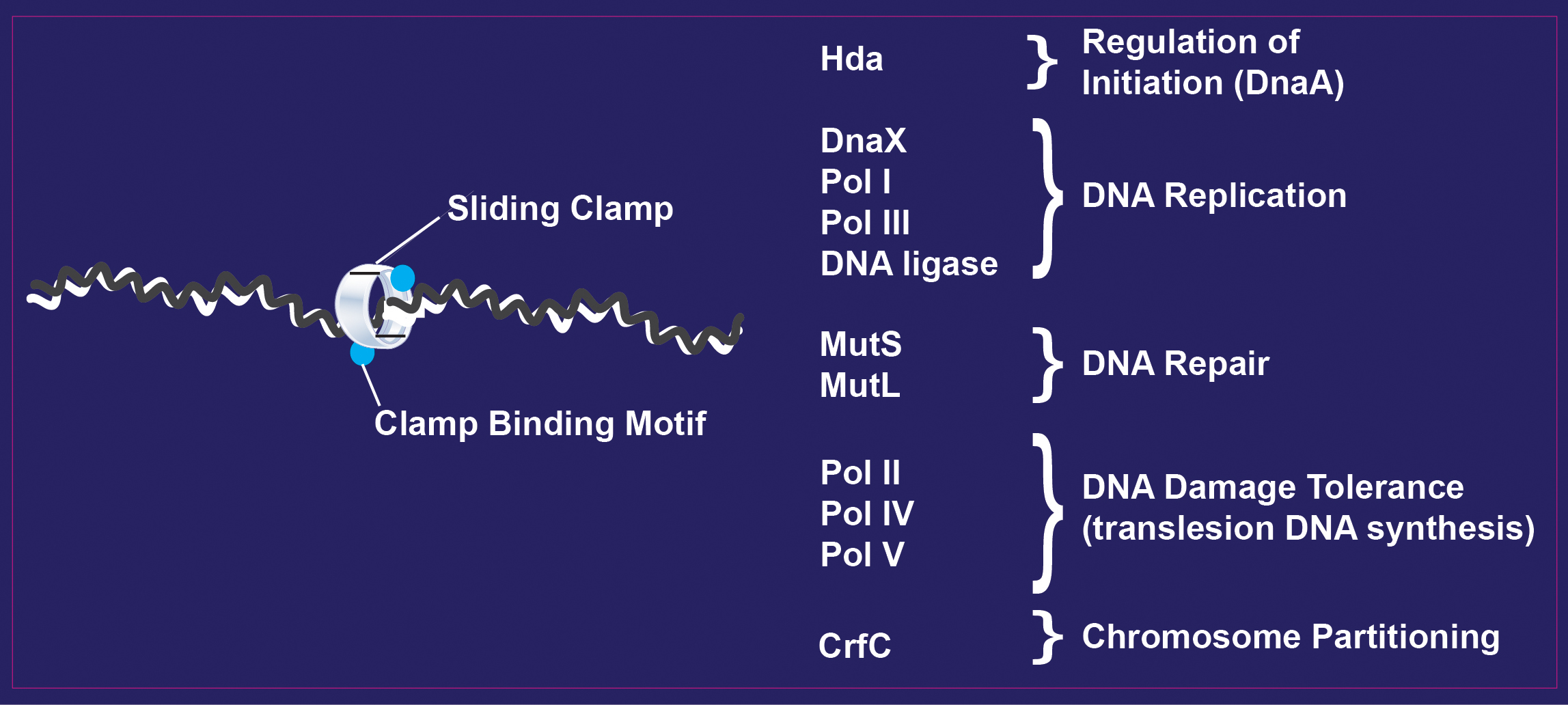
The sliding clamp: a promiscuous DNA replication protein
The term “faithful” is often used in describing DNA replication by DNA polymerases. After all, organisms do not want errors in their DNA when replisomes duplicate chromosomes. A key component of the replisome is the sliding clamp (beta clamp in bacteria, RFC in eukaryotes), which has a ring-like structure that encircles DNA. There, it can connect with many partner proteins to perform specific functions on that DNA. However, a molecular understanding is lacking as to both how the clamp interacts with these different partners, and the mechanisms by which it manages their respective actions.
Because of their shared interests, Drs. Mark Sutton, and Jon Kaguni collaborated on a project to study the biochemistry of the beta clamp. Dr. Sutton, a former grad student from Kaguni’s lab, is a professor in the Department of Biochemistry, Jacobs School of Medicine and Biomedical Sciences at the University at Buffalo, SUNY. His laboratory seeks to develop an integrated mechanistic view of how organisms coordinate the actions of their DNA replication machinery with those of other cellular factors involved in DNA repair and damage tolerance. Failure to properly coordinate these functions leads to mutations, genome instability, and in extreme cases, cell death.
The Kaguni lab studies the biochemistry of replication initiation, and how this process is regulated to ensure that it occurs only once per cell cycle. One mechanism of regulation is through Hda and the dimeric beta clamp, which form a complex that stimulates the hydrolysis of ATP bound to DnaA. The resulting DnaA-ADP is not active in replication initiation.
Using a variety of methods, which include structural modeling, biochemical assays, biosensor analysis, bacteriological, genetic, molecular and immunochemical methods, transcriptome analysis, flow cytometry and mutational analysis, the Sutton and Kaguni labs performed experiments that led to a pair of publications “The Mutant β E202K Sliding Clamp Protein Impairs DNA Polymerase III Replication Activity” and “Elevated Levels of the Escherichia coli nrdAB-Encoded Ribonucleotide Reductase Counteract the Toxicity Caused by an Increased Abundance of the β Clamp”.
The evidence suggests that the physical and functional interactions of the clamp with DNA polymerase III of E. coli are not limited to the clamp binding motif but are more extensive than currently appreciated. Other findings suggest that Hda, which forms a complex with the beta clamp to regulate the activity of DnaA by stimulating its intrinsic ATPase activity, also plays a noncatalytic role in regulating DnaA-ATP by sequestering it. These and other studies reveal the role of critical proteins in specific pathways that exploit the DNA-bound state of the sliding clamp.
“Despite the sometimes tough challenges raised by the various experimental assays and the obstacle of SARS-Cov2, completing the work to share with our scientific colleagues is very gratifying,” said Kaguni, a professor of biochemistry and molecular biology in MSU’s College of Natural Science.
Banner Image: The sliding clamp, a promiscuous DNA replication protein that interacts with partners via its clamp binding motif. Image: Jon Kaguni



